Even a superficial examination of the microbial world shows that bacteria are one of the most important groups by any criterion: numbers of organisms, general ecological importance, or practical importance for humans. Indeed, much of our understanding of phenomena in biochemistry and molecular biology comes from research on bacteria. Although considerable space is devoted to eucaryotic microorganisms, the major focus is on procaryotes.
Therefore
the unit on microbial morphology begins with the structure of procaryotes. There
are two quite different groups of procaryotes: Bacteria and Archaea. This
chapter focuses primarily on bacterial morphology; chapter 20 will discuss
archaeal cell structure and composition. A comment about nomenclature is
necessary to avoid confusion. The word procaryote will be used in a general
sense to include both the bacteria and archaea; the term bacterium will refer
specifically to bacteria. Eucaryotes, procaryotes, and the composition of the
microbial world.
3.1 An Overview of Bacterial Cell Structure
Because
much of this chapter is devoted to a discussion of individual cell components,
a preliminary overview of the prokaryotic cell as a whole is in order.
Size, Shape, and Arrangement of Bacteria
One
might expect that small, relatively simple organisms like procaryotes would be
uniform in shape and size. Although it is true that many procaryotes are similar
in morphology, there is a remarkable amount of variation due to differences in
genetics and ecology. (figures 3.1 and 3.2; see also figures
2.8 and 2.15).
Major
morphological patterns are described here, and interesting variants are
mentioned in the procaryotic survey.
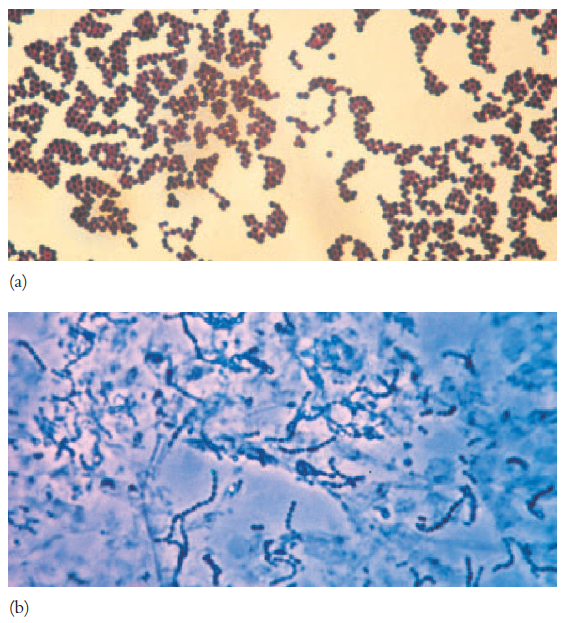
Most
commonly encountered bacteria have one of two shapes. Cocci (s.,coccus)
are roughly spherical cells. They can exist as individual cells, but also are
associated in characteristic arrangements that are frequently useful in
bacterial identification.
Diplococci (s., diplococcus) arise when cocci
divide and remain together to form pairs (Neisseria; see figure 2.15d). Long
chains of cocci result when cells adhere after repeated divisions in one plane;
this pattern is seen in the genera Streptococcus, Enterococcus, and Lactococcus
(figure 3.1b). Staphylococcus divides in random planes to
generate irregular grapelike clumps (figure 3.1a). Divisions in two or
three planes can produce symmetrical clusters of cocci. Members of the genus Micrococcus
often divide in two planes to form square groups of four cells called
tetrads. In the genus Sarcina, cocci divide in three planes producing
cubical packets of eight cells.
The
other common bacterial shape is that of a rod, often called a bacillus
(pl., bacilli). Bacillus megaterium is a typical example of a
bacterium with a rod shape (figure 3.1c; see also figure 2.15a,c).
Bacilli differ considerably in their length-to width ratio, the coccobacilli
being so short and wide that they resemble cocci. The shape of the rod’s end
often varies between species and may be flat, rounded, cigar-shaped, or
bifurcated.
Although
many rods do occur singly, they may remain together after division to form
pairs or chains (e.g., Bacillus megaterium is found in long chains). A
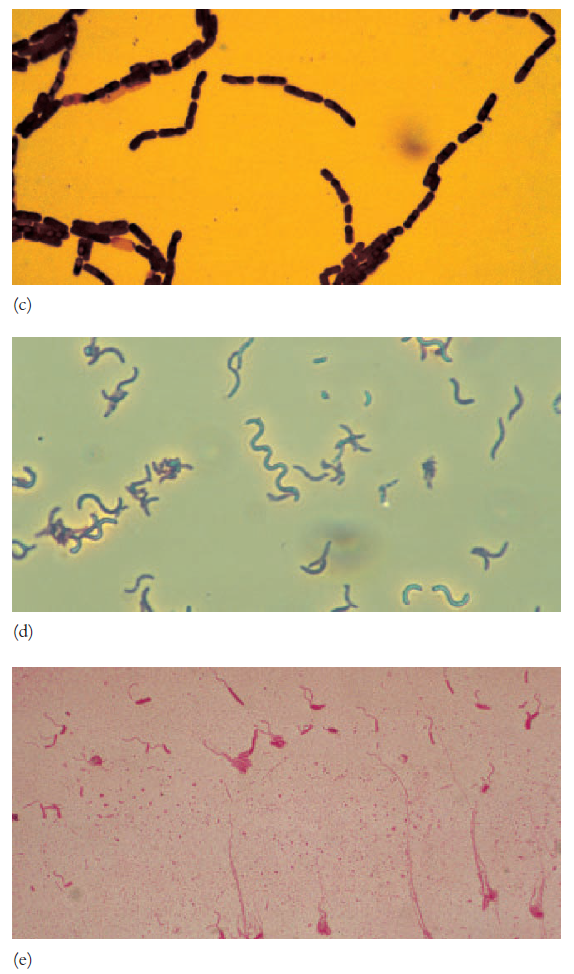
few rod-shaped bacteria, the vibrios, are curved to form distinctive
commas or incomplete spirals (figure 3.1e).
Bacteria
can assume a great variety of shapes, although they often are simple spheres or
rods. Actinomycetes characteristically form long multinucleate filaments or
hyphae that may branch to produce a network called a mycelium (figure 3.2a).
Many bacteria are shaped like long rods twisted into spirals or helices; they
are called spirilla if rigid and spirochetes when flexible
(figures 3.1d, 3.2c; see also figure 2.8a,c). The oval- to
pear-shaped Hyphomicrobium (figure 3.2d) produces a bud at the
end of a long hypha. Other bacteria such as Gallionella produce
nonliving stalks (figure 3.2f).
A few
bacteria actually are flat. For example, Anthony E. Walsby has discovered
square bacteria living in salt ponds (figure 3.2e). These bacteria are
shaped like flat, square-to rectangular boxes about 2 _m by 2 to 4 _m, and only
0.25 _m thick. Finally, some bacteria are variable in shape and lack a single,
characteristic form (figure 3.2b). These are called pleomorphic even
though they may, like Corynebacterium, have a generally rod-like form.
Bacteria vary in size as much as in shape (figure 3.3). The smallest (e.g., some members of the genus Mycoplasma) are about 0.3 µm in diameter, approximately the size of the largest viruses (the poxviruses). Recently there have been reports of even smaller cells. Nanobacteria or ultra-microbacteria appear to range from around 0.2 µm to less than 0.05 µm in diameter.
A few strains have been cultured, but most are simply very small bacteria-like objects only observed microscopically. It has been thought that the smallest possible cell is about 0.14 to 0.2 µm in diameter, but many nanobacteria are reported to be smaller. Some microbiologists think nanobacteria are artifacts, and more research will be required before the significance of these forms becomes clear. Escherichia coli, a bacillus of about average size, is 1.1 to 1.5 µm wide by 2.0 to 6.0 µm long. A few bacteria become fairly large; some spirochetes occasionally reach 500 µm in length, and the cyanobacterium Oscillatoria is about 7 _m in diameter (the same diameter as a red blood cell). A huge bacterium lives in the intestine of the brown surgeonfish, Acanthurus nigrofuscus. Epulopiscium fishelsoni grows as large as 600 by 80 µm, a little smaller than a printed hyphen. More recently an even larger bacterium, Thiomargarita namibiensis, has been discovered in ocean sediment (Box 3.1). Thus a few bacteria are much larger than the average eucaryotic cell (typical plant and animal cells are around 10–50 µm in diameter).
Bacterial Cell Organization
A
variety of structures is found in procaryotic cells. Their major functions are
summarized in table 3.1, and figure 3.4 illustrates many of them. Not all
structures are found in every genus. Furthermore, gram-negative and
gram-positive cells differ, particularly with respect to their cell walls.
Despite these variations procaryotes are consistent in their fundamental
structure and most important components.
Procaryotic
cells almost always are bounded by a chemically complex cell wall. Inside this
wall, and separated from it by a periplasmic space, lies the plasma membrane.
This membrane can be invaginated to form simple internal membranous structures.
Since the procaryotic cell does not contain internal membrane-bound organelles,
its interior appears morphologically simple. The genetic material is localized
in a discrete region, the nucleoid, and is not separated from the surrounding
cytoplasm by membranes. Ribosomes and larger masses called inclusion bodies are
scattered about in the cytoplasmic matrix. Both gram-positive and gram-negative
cells can use flagella for locomotion. In addition, many cells are surrounded by
a capsule or slime layer external to the cell wall.
Procaryotic
cells are morphologically much simpler than eukaryotic cells. These two cell types
are compared following the review of eucaryotic cell structure.
3.2 Bacterial Cell Membranes
Membranes
are an absolute requirement for all living organisms. Cells must interact in a
selective fashion with their environment, whether it is the internal
environment of a multicellular organism or a less protected and more variable
external environment. Cells must not only be able to acquire nutrients and
eliminate wastes, but they also have to maintain their interior in a constant,
highly organized state in the face of external changes. The plasma membrane
encompasses the cytoplasm of both procaryotic and eucaryotic cells. This
membrane is the chief point of contact with the cell’s environment and thus is
responsible for much of its relationship with the outside world. To understand
membrane function, it is necessary to become familiar with membrane structure,
and particularly with plasma membrane structure.
The Bacterial Plasma Membrane
Membranes
contain both proteins and lipids, although the exact proportions of protein and
lipid vary widely. Bacterial plasma membranes usually have a higher proportion
of protein than do eucaryotic membranes, presumably because they fulfill so
many different functions that are carried out by other organelle membranes in
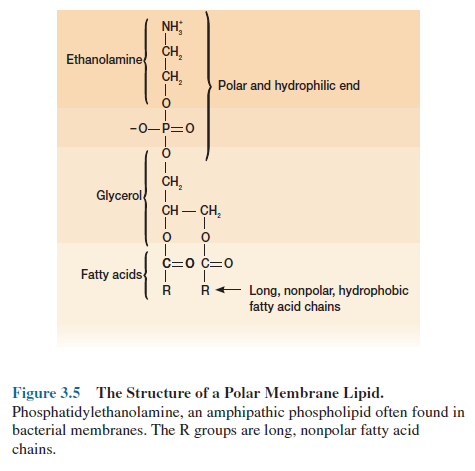
eucaryotes. Most membrane-associated lipids are structurally asymmetric with
polar and nonpolar ends (figure 3.5) and are called amphipathic. The polar ends
interact with water and are hydrophilic; the nonpolar hydrophobic ends are
insoluble in water and tend to associate with one another. This property of lipids
enables them to form a bilayer in membranes. The outer surfaces are
hydrophilic, whereas hydrophobic ends are buried in the interior away from the
surrounding water. Many of these amphipathic lipids are phospholipids (figure
3.5). Bacterial membranes usually differ from eucaryotic membranes in lacking sterols
such as cholesterol (figure 3.6a). However, many bacterial membranes do
contain penta-cyclic sterol-like molecules called hopanoids (figure 3.6b),
and huge quantities of hopanoids are present in our ecosystem (Box 3.2).
Hopanoids are synthesized from the same precursors as steroids. Like steroids
in eucaryotes, they probably stabilize the bacterial membrane. The membrane
lipid is organized in two layers, or sheets, of molecules arranged end-to-end
(figure 3.7).
Many
archaeal membranes differ from other bacterial membranes in having a monolayer
with lipid molecules spanning the whole membrane. Cell membranes are very thin
structures, about 5 to 10 nm thick, and can only be seen with the electron
microscope. The freeze-etching technique has been used to cleave membranes down
the center of the lipid bilayer, splitting them in half and exposing the
interior. In this way it has been discovered that many membranes, including the
plasma membrane, have a complex internal structure. The small globular
particles seen in these membranes are thought to be membrane proteins that lie
within the membrane lipid bilayer (see figure 2.26).
The
most widely accepted current model for membrane structure is the fluid mosaic
model of S. Jonathan Singer and Garth Nicholson (figure 3.7). They distinguish
between two types of membrane proteins. Peripheral proteins are loosely
connected to the membrane and can be easily removed. They are soluble in
aqueous solutions and make up about 20 to 30% of total membrane protein. About
70 to 80% of membrane proteins are integral proteins. These are not easily
extracted from membranes and are insoluble in aqueous solutions when freed of lipids.
Integral proteins, like membrane lipids, are amphipathic; their hydrophobic
regions are buried in the lipid while the hydrophilic portions project from the
membrane surface (figure 3.7). Some of these proteins even extend all the way
through the lipid layer. Integral proteins can diffuse laterally around the
surface to new locations, but do not flip-flop or rotate through the lipid
layer. Often carbohydrates are attached to the outer surface of plasma membrane
proteins and seem to have important functions.
The
emerging picture of the cell membrane is one of a highly organized and
asymmetric system, which also is flexible and dynamic. Although membranes
apparently have a common basic design, there are wide variations in both their
structure and functional capacities. The differences are so large and
characteristic that membrane chemistry can be used in bacterial identification.
The
plasma membranes of procaryotic cells must fill an incredible variety of roles
successfully. Many major plasma membrane functions are noted here even though
they are discussed individually at later points in the text. The plasma
membrane retains the cytoplasm, particularly in cells without cell walls, and
separates it from the surroundings. The plasma membrane also serves as a
selectively permeable barrier: it allows particular ions and molecules to pass,
either into or out of the cell, while preventing the movement of others. Thus
the membrane prevents the loss of essential components through leakage while
allowing the movement of other molecules. Because many substances cannot cross
the plasma membrane without assistance, it must aid such movement when necessary.
Transport systems can be used for such tasks as nutrient uptake, waste
excretion, and protein secretion. The prokaryotic plasma membrane also is the
location of a variety of crucial metabolic processes: respiration,
photosynthesis, the synthesis of lipids and cell wall constituents, and
probably chromosome segregation.
Finally,
the membrane contains special receptor molecules that help procaryotes detect
and respond to chemicals in their surroundings. Clearly the plasma membrane is
essential to the survival of microorganisms.
Bacterial Internal
Membrane Systems
Although
procaryotic cytoplasm does not contain complex membranous organelles like
mitochondria or chloroplasts, membranous structures of several kinds can be
observed. A commonly observed structure is the mesosome. Mesosomes are
invaginations of the plasma membrane in the shape of vesicles, tubules, or lamellae
(figure 3.8 and figure 3.11). They are seen in both gram-positive and
gram-negative bacteria, although they are generally more prominent in the
former.
Mesosomes
often are found next to septa or cross-walls in dividing bacteria and sometimes
seem attached to the bacterial chromosome. Thus they may be involved in cell
wall formation during division or play a role in chromosome replication and
distribution to daughter cells.
Currently
many bacteriologists believe that mesosomes are artifacts generated during the
chemical fixation of bacteria for electron microscopy. Possibly they represent
parts of the plasma membrane that are chemically different and more disrupted
by fixatives.
Many
bacteria have internal membrane systems quite different from the mesosome
(figure 3.9). Plasma membrane infoldings can become extensive and complex in
photosynthetic bacteria such as the cyanobacteria and purple bacteria or in
bacteria with very high respiratory activity like the nitrifying bacteria. They
may be aggregates of spherical vesicles, flattened vesicles, or tubular
membranes. Their function may be to provide a larger membrane surface for
greater metabolic activity.
3.3 The Bacterial Cytoplasmic Matrix
Procaryotic
cytoplasm, unlike that of eucaryotes, lacks unit membrane-bound organelles. The
cytoplasmic matrix is the substance lying between the plasma membrane and the
nucleoid (p. 54). The matrix is largely water (about 70% of bacterial mass is
water). It is featureless in electron micrographs but often is packed with
ribosomes and highly organized (figure 3.10). Specific proteins are positioned
at particular sites such as the cell pole and the place where the bacterial
cell will divide. Thus although bacteria may lack a true cytoskeleton, they do
have a cytoskeleton like system of proteins in their cytoplasmic matrix. The
plasma membrane and everything within is called the protoplast; thus the cytoplasmic
matrix is a major part of the protoplast.
Bacterial Inclusion
Bodies
A
variety of inclusion bodies, granules of organic or inorganic material that
often are clearly visible in a light microscope, is present in the cytoplasmic
matrix. These bodies usually are used for storage (e.g., carbon compounds,
inorganic substances, and energy), and also reduce osmotic pressure by tying up
molecules in particulate form. Some inclusion bodies are not bounded by a
membrane and lie free in the cytoplasm—for example, polyphosphate granules,
cyanophycin granules, and some glycogen granules. Other inclusion bodies are
enclosed by a membrane about 2.0 to 4.0 nm thick, which is single-layered and
not a typical bilayer membrane. Examples of membrane-enclosed inclusion bodies
are poly-_-hydroxybutyrate granules, some glycogen and sulfur granules,
carboxysomes, and gas vacuoles.
Inclusion
body membranes vary in composition. Some are protein in nature, whereas others
contain lipid. Because inclusion bodies are used for storage, their quantity
will vary with the nutritional status of the cell. For example, polyphosphate granules
will be depleted in freshwater habitats that are phosphate limited. A brief
description of several important inclusion bodies follows.
Organic
inclusion bodies usually contain either glycogen or poly-β-hydroxybutyrate.
Glycogen is a polymer of glucose units composed of long chains formed by α(1→4)
glycosidic bonds and branching chains connected to them by α (1→6)
glycosidic bonds . Poly-β-hydroxybutyrate (PHB) contains β-hydroxybutyrate
molecules joined by ester bonds between the carboxyl and hydroxyl groups of
adjacent molecules.
Usually
only one of these polymers is found in a species, but purple photosynthetic
bacteria have both. Poly-β-hydroxybutyrate accumulates in distinct bodies,
around 0.2 to 0.7 µm in diameter, that are readily stained with Sudan black for
light microscopy and are clearly visible in the electron microscope (figure
3.11).
Glycogen
is dispersed more evenly throughout the matrix as small granules (about 20 to
100 nm in diameter) and often can be seen only with the electron microscope. If
cells contain a large amount of glycogen, staining with an iodine solution will
turn them reddish-brown. Glycogen and PHB inclusion bodies are carbon storage
reservoirs providing material for energy and biosynthesis. Many bacteria also
store carbon as lipid droplets.
Cyanobacteria
have two distinctive organic inclusion bodies. Cyanophycin granules (figure
3.13a) are composed of large polypeptides containing approximately equal
amounts of the amino acids arginine and aspartic acid. The granules often are large
enough to be visible in the light microscope and store extra nitrogen for the
bacteria. Carboxysomes are present in many cyanobacteria, nitrifying bacteria,
and thiobacilli. They are polyhedral, about 100 nm in diameter, and contain the
enzyme ribulose- 1,5-bisphosphate carboxylase in a paracrystalline arrangement.
They serve as a reserve of this enzyme and may be a site of CO2 fixation.
A
most remarkable organic inclusion body, the gas vacuole, is present in many
cyanobacteria, purple and green photosynthetic bacteria, and a few other
aquatic forms such as Halobacterium and Thiothrix. These bacteria
float at or near the surface, because gas vacuoles give them buoyancy. This is
vividly demonstrated by a simple but dramatic experiment. Cyanobacteria held in
a full, tightly stoppered bottle will float, but if the stopper is struck with
a hammer, the bacteria sink to the bottom. Examination of the bacteria at the
beginning and end of the experiment shows that the sudden pressure increase has
collapsed the gas vacuoles and destroyed the microorganisms’ buoyancy.
Gas
vacuoles are aggregates of enormous numbers of small, hollow, cylindrical
structures called gas vesicles (figure 3.12).
Gas
vesicle walls do not contain lipid and are composed entirely of a single small
protein. These protein subunits assemble to form a rigid enclosed cylinder that
is hollow and impermeable to water but freely permeable to atmospheric gases.
Bacteria with gas vacuoles can regulate their buoyancy to float at the depth
necessary for proper light intensity, oxygen concentration, and nutrient
levels.
They
descend by simply collapsing vesicles and float upward when new ones are
constructed.
Two
major types of inorganic inclusion bodies are seen. Many bacteria store
phosphate as polyphosphate granules or volutin granules (figure 3.13a).
Polyphosphate is a linear polymer of orthophosphates joined by ester bonds.
Thus volutin granules function as storage reservoirs for phosphate, an
important component of cell constituents such as nucleic acids. In some cells
they act as an energy reserve, and polyphosphate can serve as an energy source
in reactions. These granules are sometimes called metachromatic granules
because they show the metachromatic effect; that is, they appear red or a different
shade of blue when stained with the blue dyes methylene blue or toluidine blue.
Some bacteria also store sulfur temporarily as sulfur granules, a second type
of inorganic inclusion body (figure 3.13b).
For
example, purple photosynthetic bacteria can use hydrogen sulfide as a
photosynthetic electron donor and accumulate the resulting sulfur in either the
periplasmic space or in special cytoplasmic globules.
Inorganic
inclusion bodies can be used for purposes other than storage. An excellent
example is the magnetosome, which is used by some bacteria to orient in the
earth’s magnetic field. These inclusion bodies contain iron in the form of
magnetite (Box 3.3).
Bacterial Ribosomes
As mentioned earlier, the cytoplasmic matrix often is packed with ribosomes; they also may be loosely attached to the plasma membrane. Ribosomes look like small, featureless particles at low magnification in electron micrographs (figure 3.11) but are actually very complex objects made of both protein and ribonucleic acid (RNA). They are the site of protein synthesis; matrix ribosomes synthesize proteins destined to remain within the cell, whereas the plasma membrane ribosomes make proteins for transport to the outside. The newly formed polypeptide folds into its final shape either as it is synthesized by the ribosome or shortly after completion of protein synthesis. The shape of each protein is determined by its amino acid sequence.
Special proteins called molecular
chaperones, or chaperones, aid the polypeptide in folding to its proper shape.
Note that procaryotic ribosomes are smaller than eukaryotic ribosomes. They
commonly are called 70S ribosomes, have dimensions of about 14 to 15 nm by 20
nm, a molecular weight of approximately 2.7 million, and are constructed of a
50S and a 30S subunit. The S in 70S and similar values stands for Svedberg unit.
This is the unit of the sedimentation coefficient, a measure of the
sedimentation velocity in a centrifuge; the faster a particle travels when
centrifuged, the greater its Svedberg value or sedimentation coefficient. The
sedimentation coefficient is a function of a particle’s molecular weight,
volume, and shape (see figure 16.7). Heavier and more compact particles
normally have larger Svedberg numbers or sediment faster. Ribosomes in the
cytoplasmic matrix of eucaryotic cells are 80S ribosomes and about 22 nm in
diameter. Despite their overall difference in size, both types of ribosomes are
similarly composed of a large and a small subunit.
3.4 The Bacterial Nucleoid
Probably the most striking
difference between procaryotes and eucaryotes is the way in which their genetic
material is packaged. Eucaryotic cells have two or more chromosomes contained within
a membrane-delimited organelle, the nucleus. In contrast, procaryotes lack a
membrane-delimited nucleus. The prokaryotic chromosome is located in an
irregularly shaped region called the nucleoid (other names are also used: the
nuclear body, chromatin body, nuclear region). Usually procaryotes contain a
single circle of double-stranded deoxyribonucleic acid (DNA), but some have a
linear DNA chromosome. Recently it has been discovered that some bacteria such
as Vibrio cholerae have more than one chromosome. Although nucleoid
appearance varies with the method of fixation and staining, fibers often are
seen in electron micrographs (figure 3.11 and figure 3.14) and are probably
DNA.
The nucleoid also is
visible in the light microscope after staining with the Feulgen stain, which
specifically reacts with DNA. A cell can have more than one nucleoid when cell
division occurs after the genetic material has been duplicated (figure 3.14a).
In actively growing bacteria, the nucleoid has projections that extend into the
cytoplasmic matrix (figure 3.14b,c). Presumably these projections contain
DNA that is being actively transcribed to produce mRNA.
Careful electron microscopic
studies often have shown the nucleoid in contact with either the mesosome or
the plasma membrane. Membranes also are found attached to isolated nucleoids.
Thus there is evidence that
bacterial DNA is attached to cell membranes, and membranes may be involved in
the separation of DNA into daughter cells during division.
Nucleoids have been
isolated intact and free from membranes. Chemical analysis reveals that they
are composed of about 60% DNA, 30% RNA, and 10% protein by weight. In Escherichia
coli, a rod-shaped cell about 2 to 6 µm long, the closed DNA circle
measures approximately 1,400 µm. Obviously it must be very efficiently packaged
to fit within the nucleoid.
The DNA is looped and
coiled extensively (see figure 11.8), probably with the aid of RNA and
nucleoid proteins (these proteins differ from the histone proteins present in eukaryotic
nuclei).
There are a few exceptions
to the above picture. Membrane-bound DNA-containing regions are present in two
genera of planctomycetes. Pirellula has a single membrane that surrounds
a region, the pirellulosome, which contains a fibrillar nucleoid and
ribosome-like particles. The nuclear body of Gemmata obscuriglobus is
bounded by two membranes (see figure 21.12).
More work will be required
to determine the functions of these membranes and how widespread this
phenomenon is. Many bacteria possess plasmids in addition to their chromosome. These
are double-stranded DNA molecules, usually circular, that can exist and
replicate independently of the chromosome or may be integrated with it; in
either case they normally are inherited or passed on to the progeny. However,
plasmids are not usually attached to the plasma membrane and sometimes are lost
to one of the progeny cells during division. Plasmids are not required for host
growth and reproduction, although they may carry genes that give their
bacterial host a selective advantage. Plasmid genes can render bacteria
drug-resistant, give them new metabolic abilities, make them pathogenic, or
endow them with a number of other properties. Because plasmids often move
between bacteria, properties such as drug resistance can spread throughout a
population.
3.5 The Bacterial Cell Wall
The cell wall is the
layer, usually fairly rigid, that lies just outside the plasma membrane. It is
one of the most important parts of a procaryotic cell for several reasons.
Except for the mycoplasmas and some Archaea, most bacteria have strong walls
that give them shape and protect them from osmotic lysis wall shape and
strength is primarily due to peptidoglycan, as we will see shortly. The cell
walls of many pathogens have components that contribute to their pathogenicity.
The wall can protect a
cell from toxic substances and is the site of action of several antibiotics.
After Christian Gram developed
the Gram stain in 1884, it soon became evident that bacteria could be divided
into two major groups based on their response to the Gram-stain procedure.
Gram-positive bacteria stained purple, whereas gram-negative bacteria were
colored pink or red by the technique.
The true structural
difference between these two groups became clear with the advent of the
transmission electron microscope.
The gram-positive cell
wall consists of a single 20 to 80 nm thick homogeneous peptidoglycan or murein
layer lying outside the plasma membrane (figure 3.15). In contrast, the
gram-negative cell wall is quite complex. It has a 2 to 7 nm peptidoglycan
layer surrounded by a 7 to 8 nm thick outer membrane. Because of the thicker
peptidoglycan layer, the walls of gram-positive cells are stronger than those
of gram-negative bacteria. Microbiologists often call all the structures from
the plasma membrane outward the envelope or cell envelope. This includes the
wall and structures like capsules when present.
Frequently a space is seen between the plasma membrane and the outer membrane in electron micrographs of gram-negative bacteria, and sometimes a similar but smaller gap may be observed between the plasma membrane and wall in gram-positive bacteria. This space is called the periplasmic space. Recent evidence indicates that the periplasmic space may be filled with a loose network of peptidoglycan. Possibly it is more a gel than a fluid-filled space. The substance that occupies the periplasmic space is the periplasm.
Gram-positive cells may have periplasm even if they lack a discrete, obvious periplasmic space. Size estimates of the periplasmic space in gram-negative bacteria range from 1 nm to as great as 71 nm. Some recent studies indicate that it may constitute about 20 to 40% of the total cell volume (around 30 to 70 nm), but more research is required to establish an accurate value. When cell walls are disrupted carefully or removed without disturbing the underlying plasma membrane, periplasmic enzymes and other proteins are released and may be easily studied. The periplasmic space of gram-negative bacteria contains many proteins that participate in nutrient acquisition— for example, hydrolytic enzymes attacking nucleic acids and phosphorylated molecules, and binding proteins involved in transport of materials into the cell.
Denitrifying and chemolithoautotrophic bacteria often have electron transport
proteins in their periplasm. The periplasmic space also contains enzymes
involved in peptidoglycan synthesis and the modification of toxic compounds
that could harm the cell. Gram-positive bacteria may not have a visible
periplasmic space and do not appear to have as many periplasmic proteins;
rather, they secrete several enzymes that ordinarily would be periplasmic in gram-negative
bacteria. Such secreted enzymes are often called exoenzymes. Some enzymes
remain in the periplasm and are attached to the plasma membrane.
The Archaea differ from
other procaryotes in many respects. Although they may be either gram positive
or gram negative, their cell walls are distinctive in structure and chemical
composition. The walls lack peptidoglycan and are composed of proteins,
glycoproteins, or polysaccharides.
Following this overview of
the envelope, peptidoglycan structure and the organization of gram-positive and
gram-negative cell walls are discussed in more detail.
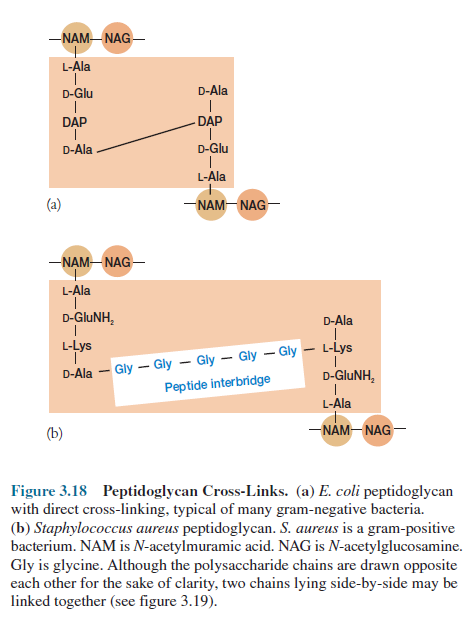
Bacterial Peptidoglycan
Structure
Peptidoglycan or murein is
an enormous polymer composed of many identical subunits. The polymer contains
two sugar derivatives, N-acetylglucosamine and N-acetylmuramic
acid (the lactyl ether of N-acetylglucosamine), and several different
amino acids, three of which—D-glutamic acid, D-alanine, and meso-diaminopimelic
acid—are not found in proteins. The presence of D-amino acids protects against
attack by most peptidases. The peptidoglycan subunit present in most
gram-negative bacteria and many gram-positive ones is shown in figure 3.16. The
backbone of this polymer is composed of alternating N-acetylglucosamine
and N-acetylmuramic acid residues. A peptide chain of four alternating
D- and L-amino acids is connected to the carboxyl group of N-acetylmuramic
acid. Many bacteria substitute another diaminoacid, usually L-lysine, in the
third position for meso-diaminopimelic acid (figure 3.17).
Chains of linked
peptidoglycan subunits are joined by cross-links between the peptides. Often
the carboxyl group of the terminal D-alanine is connected directly to the amino
group of diaminopimelic acid, but a peptide interbridge may be used instead
(figure 3.18). Most gram-negative cell wall peptidoglycan lacks the peptide
interbridge. This cross-linking results in an enormous peptidoglycan sac that
is actually one dense, interconnected network (figure 3.19). These sacs have
been isolated from gram-positive bacteria and are strong enough to retain their
shape and integrity (figure 3.20), yet they are elastic and somewhat stretchable,
unlike cellulose. They also must be porous, as molecules can penetrate them.
Bacterial Gram-Positive Cell Walls
Normally the thick, homogeneous cell
wall of gram-positive bacteria is composed primarily of peptidoglycan, which
often contains a peptide interbridge (figure 3.20 and figure 3.21). However gram-positive
cell walls usually also contain large amounts of teichoic acids, polymers of glycerol
or ribitol joined by phosphate groups (figures 3.21 and 3.22). Amino acids such
as D-alanine or sugars like glucose are attached to the glycerol and ribitol
groups.
The teichoic acids are connected to either
the peptidoglycan itself by a covalent bond with the six hydroxyl of N-acetylmuramic
acid or to plasma membrane lipids; in the latter case they are called
lipoteichoic acids. Teichoic acids appear to extend to the surface of the
peptidoglycan, and, because they are negatively charged, help give the
gram-positive cell wall its negative charge.
The functions of these molecules are
still unclear, but they may be important in maintaining the structure of the
wall. Teichoic acids are not present in gram-negative bacteria.
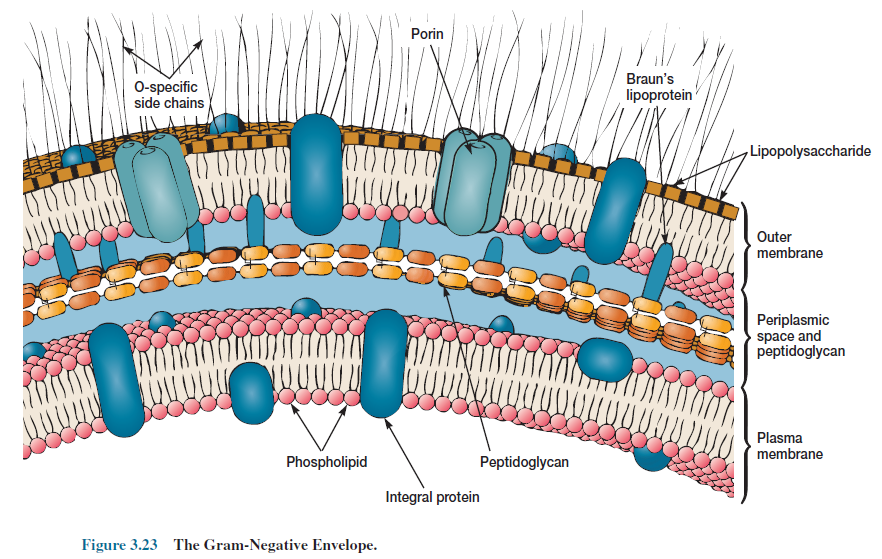
Bacterial Gram-Negative Cell Walls
Even a brief inspection of figure 3.15
shows that gram-negative cell walls are much more complex than gram-positive
walls. The thin peptidoglycan layer next to the plasma membrane may constitute
not more than 5 to 10% of the wall weight. In E. coli it is about 2 nm
thick and contains only one or two layers or sheets of peptidoglycan.
The outer membrane lies outside the
thin peptidoglycan layer (figures 3.23 and 3.24). The most abundant membrane
protein is Braun’s lipoprotein, a small lipoprotein covalently joined to the
underlying peptidoglycan and embedded in the outer membrane by its hydrophobic
end. The outer membrane and peptidoglycan are so firmly linked by this
lipoprotein that they can be isolated as one unit.
Another structure that may strengthen
the gram-negative wall and hold the outer membrane in place is the adhesion
site.
The outer membrane and plasma membrane
appear to be in direct contact at many locations in the gram-negative wall. In E.
coli 20 to 100 nm areas of contact between the two membranes are seen in
plasmolyzed cells. Adhesion sites may be regions of direct contact or possibly true
membrane fusions. It has been proposed that substances can move into the cell
through these adhesion sites rather than traveling through the periplasm.
Possibly the most unusual constituents
of the outer membrane are its lipopolysaccharides (LPSs). These large, complex molecules
contain both lipid and carbohydrate, and consist of three parts: (1) lipid A,
(2) the core polysaccharide, and (3) the O side chain. The LPS from Salmonella
typhimurium has been studied most, and its general structure is described
here (figure 3.25). The lipid A region contains two glucosamine sugar
derivatives, each with three fatty acids and phosphate or pyrophosphate
attached. It is buried in the outer membrane and the remainder of the LPS
molecule projects from the surface.
The core polysaccharide is joined to
lipid A. In Salmonella it is constructed of 10 sugars, many of them
unusual in structure. The O side chain or O antigen is a polysaccharide chain
extending outward from the core. It has several peculiar sugars and varies in
composition between bacterial strains. Although O side chains are readily
recognized by host antibodies, gram-negative bacteria may thwart host defenses
by rapidly changing the nature of their O side chains to avoid detection.
Antibody interaction with the LPS before reaching the outer membrane proper may
also protect the cell wall from direct attack.
The LPS is important for several reasons
other than the avoidance of host defenses. Since the core polysaccharide
usually contains charged sugars and phosphate (figure 3.25), LPS contributes to
the negative charge on the bacterial surface. Lipid A is a major constituent of
the outer membrane, and the LPS helps stabilize membrane structure.
Furthermore, lipid A often is toxic; as a result the LPS can act as an
endotoxin and cause some of the symptoms that arise in gram-negative bacterial
infections.
A most important outer membrane function
is to serve as a protective barrier. It prevents or slows the entry of bile
salts, antibiotics, and other toxic substances that might kill or injure the
bacterium.
Even so, the outer membrane is more
permeable than the plasma membrane and permits the passage of small molecules
like glucose and other monosaccharides. This is due to the presence of special porin
proteins (figures 3.23 and 3.24). Three porin molecules cluster together and
span the outer membrane to form a narrow channel through which molecules
smaller than about 600 to 700 daltons can pass. Larger molecules such as
vitamin B12 must be transported across the outer membrane by specific carriers.
The outer membrane also prevents the loss of constituents like periplasmic
enzymes.
The Mechanism of Gram Staining
Although several explanations have been given for the Gram stain reaction results, it seems likely that the difference between gram-positive and gram-negative bacteria is due to the physical nature of their cell walls. If the cell wall is removed from gram-positive bacteria, they become gram negative.
The peptidoglycan itself is not stained; instead it seems to act as a permeability barrier preventing loss of crystal violet. During the procedure the bacteria are first stained with crystal violet and next treated with iodine to promote dye retention. When gram-positive bacteria then are decolorized with ethanol, the alcohol is thought to shrink the pores of the thick peptidoglycan. Thus the dye-iodine complex is retained during the short decolorization step and the bacteria remain purple.
In
contrast, gram-negative peptidoglycan is very thin, not as highly cross-linked,
and has larger pores. Alcohol treatment also may extract enough lipid from the
gram-negative wall to increase its porosity further. For these reasons, alcohol
more readily removes the purple crystal violet-iodine complex from gram-negative
bacteria.
The Cell Wall
and Osmotic Protection
The cell wall usually is required to protect bacteria against destruction by osmotic pressure. Solutes are much more concentrated in bacterial cytoplasm than in most microbial habitats, which are hypotonic. During osmosis, water moves across selectively permeable membranes such as the plasma membrane from dilute solutions (higher water concentration) to more concentrated solutions (lower water concentration).
Thus water normally enters bacterial cells and the
osmotic pressure may reach 20 atmospheres or 300 pounds/square inch. The plasma
membrane cannot withstand such pressures and the cell will swell and be physically
disrupted and destroyed, a process called lysis, without the wall that resists
cell swelling and protects it. Solutes are more concentrated in hypertonic
habitats than in the cell. Thus water flows outward, and the cytoplasm shrivels
up and pulls away from the cell wall. This phenomenon is known as plasmolysis and
is useful in food preservation because many microorganisms cannot grow in dried
foods and jellies as they cannot avoid plasmolysis.
The
importance of the cell wall in protecting bacteria against osmotic lysis is
demonstrated by treatment with lysozyme or penicillin. The enzyme lysozyme
attacks peptidoglycan by hydrolyzing the bond that connects N-acetylmuramic
acid with carbon four of N-acetylglucosamine. Penicillin inhibits
peptidoglycan synthesis. If bacteria are incubated with penicillin in an
isotonic solution, gram-positive bacteria are converted to protoplasts that
continue to grow normally when isotonicity is maintained even though they
completely lack a wall.
Gram-negative
cells retain their outer membrane after penicillin treatment and are classified
as spheroplasts because some of their cell wall remains. Protoplasts and
spheroplasts are osmotically sensitive. If they are transferred to a dilute
solution, they will lyse due to uncontrolled water influx (figure 3.26).
Although
most bacteria require an intact cell wall for survival, some have none at all.
For example, the mycoplasmas lack a cell wall and are osmotically sensitive,
yet often can grow in dilute media or terrestrial environments because their plasma
membranes are stronger than normal. The precise reason for this is not known,
although the presence of sterols in the membranes of many species may provide
added strength. Without a rigid cell wall, mycoplasmas tend to be pleomorphic
or variable in shape.
3.6
Components External to the Bacterial Cell Wall
Bacteria
have a variety of structures outside the cell wall that can function in
protection, attachment to objects, and cell movement. Several of these are
discussed.
Capsules,
Slime Layers, and S-Layers
Some
bacteria have a layer of material lying outside the cell wall. When the layer
is well organized and not easily washed off, it is called a capsule. A slime
layer is a zone of diffuse, unorganized material that is removed easily. A
glycocalyx (figure 3.27) is a network of polysaccharides extending from the
surface of bacteria and other cells (in this sense it could encompass both
capsules and slime layers). Capsules and slime layers usually are composed of
polysaccharides, but they may be constructed of other materials. For example, Bacillus
anthracis has a capsule of poly-D-glutamic acid. Capsules are clearly
visible in the light microscope when negative stains or special capsule stains
are employed (figure 3.27a); they also can be studied with the electron
microscope (figure 3.27b).
Although capsules are not required for bacterial growth and reproduction in laboratory cultures, they do confer several advantages when bacteria grow in their normal habitats. They help bacteria resist phagocytosis by host phagocytic cells. Streptococcus pneumoniae provides a classic example. When it lacks a capsule, it is destroyed easily and does not cause disease, whereas the capsulated variant quickly kills mice.
Capsules contain a great deal of water and can protect
bacteria against desiccation. They exclude bacterial viruses and most
hydrophobic toxic materials such as detergents. The glycocalyx also aids
bacterial attachment to surfaces of solid objects in aquatic environments or to
tissue surfaces in plant and animal hosts (figure 3.28). Gliding bacteria often
produce slime, which presumably aids in their motility (see Box 21.1).
Many gram-positive and gram-negative bacteria have a regularly structured layer called an S-layer on their surface. Slayers also are very common among Archaea, where they may be the only wall structure outside the plasma membrane. The Slayer has a pattern something like floor tiles and is composed of protein or glycoprotein (figure 3.29).
In gram-negative bacteria the S-layer adheres
directly to the outer membrane; it is associated with the peptidoglycan surface
in gram-positive bacteria. It may protect the cell against ion and pH
fluctuations, osmotic stress, enzymes, or the predacious bacterium Bdellovibrio.
The S-layer also helps maintain the shape and envelope rigidity of at least some
bacterial cells. It can promote cell adhesion to surfaces. Finally, the layer
seems to protect some pathogens against complement attack and phagocytosis,
thus contributing to their virulence.
Bacterial Pili
and Fimbriae
Many
gram-negative bacteria have short, fine, hair-like appendages that are thinner
than flagella and not involved in motility. These are usually called fimbriae
(s., fimbria). Although a cell may be covered with up to 1,000 fimbriae, they
are only visible in an electron microscope due to their small size (figure
3.30). They seem to be slender tubes composed of helically arranged protein
subunits and are about 3 to 10 nm in diameter and up to several micrometers long.
At least some types of fimbriae attach bacteria to solid surfaces such as rocks
in streams and host tissues.
Sex pili (s., pilus) are similar appendages,
about 1 to 10 per cell, that differ from fimbriae in the following ways. Pili
often are larger than fimbriae (around 9 to 10 nm in diameter). They are
genetically determined by sex factors or conjugative plasmids and are required
for bacterial mating. Some bacterial viruses attach specifically to receptors
on sex pili at the start of their reproductive cycle.
Bacterial Flagella and Motility
Most
motile bacteria move by use of flagella (s., flagellum), threadlike
locomotor appendages extending outward from the plasma membrane and cell wall.
They are slender, rigid structures, about 20 nm across and up to 15 or 20 µm
long. Flagella are so thin they cannot be observed directly with a bright-field
microscope, but must be stained with special techniques designed to increase their
thickness. The detailed structure of a flagellum can only be seen in the
electron microscope (figure 3.30).
Bacterial
species often differ distinctively in their patterns of flagella distribution. Monotrichous
bacteria (trichous means hair) have one flagellum; if it is located
at an end, it is said to be a polar flagellum (figure 3.31a).
Amphitrichous bacteria (amphi means “on both sides”) have a
single flagellum at each pole.
In
contrast, lophotrichous bacteria (lopho means tuft) have a cluster
of flagella at one or both ends (figure 3.31b). Flagella are spread
fairly evenly over the whole surface of peritrichous (peri means
“around”) bacteria (figure 3.31c). Flagellation patterns are very useful
in identifying bacteria.
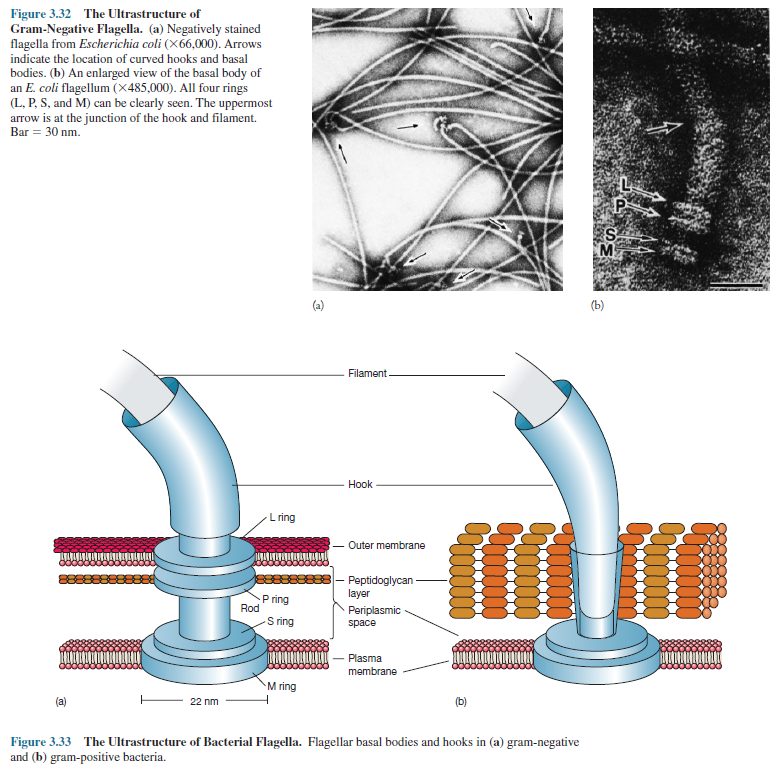
Bacterial Flagellar
Ultrastructure
Transmission
electron microscope studies have shown that the bacterial flagellum is composed
of three parts. (1) The longest and most obvious portion is the filament, which
extends from the cell surface to the tip. (2) A basal body is embedded
in the cell; and (3) a short, curved segment, the hook, links the
filament to its basal body and acts as a flexible coupling. The filament is a
hollow, rigid cylinder constructed of a single protein called flagellin, which
ranges in molecular weight from 30,000 to 60,000. The filament ends with a
capping protein. Some bacteria have sheaths surrounding their flagella. For
example Bdellovibrio has a membranous structure surrounding the
filament. Vibrio cholerae has a lipopolysaccharide sheath.
The
hook and basal body are quite different from the filament (figure 3.32).
Slightly wider than the filament, the hook is made of different protein
subunits. The basal body is the most complex part of a flagellum (figure 3.32
and figure 3.33). In E. coli and most gram-negative bacteria, the
body has four rings connected to a central rod. The outer L and P rings
associate with the lipopolysaccharide and peptidoglycan layers, respectively.
The
inner M ring contacts the plasma membrane. Gram-positive bacteria have only two
basal body rings, an inner ring connected to the plasma membrane and an outer
one probably attached to the peptidoglycan.
Bacterial Flagellar
Synthesis
The
synthesis of flagella is a complex process involving at least 20 to 30 genes.
Besides the gene for flagellin, 10 or more genes code for hook and basal body
proteins; other genes are concerned with the control of flagellar construction
or function. It is not known how the cell regulates or determines the exact
location of flagella.
Bacteria
can be deflagellated, and the regeneration of the flagellar filament can then
be studied. It is believed that flagellin subunits are transported through the
filament’s hollow internal core.
When
they reach the tip, the subunits spontaneously aggregate under the direction of
a special filament cap so that the filament grows at its tip rather than at the
base (figure 3.34). Filament synthesis is an excellent example of self-assembly.
Many structures form pontaneously through
the association of their component parts without the aid of any special enzymes
or other factors. The information required for filament construction is present
in the structure of the flagellin subunit itself.
The Mechanism
of Bacterial Flagellar Movement
Procaryotic
flagella operate differently from eucaryotic flagella. The filament is in the
shape of a rigid helix, and the bacterium moves when this helix rotates.
Considerable evidence shows that flagella act just like propellers on a boat.
Bacterial mutants with straight flagella or abnormally long hook regions
(polyhook mutants) cannot swim. When bacteria are tethered to a glass slide us-
ing antibodies to filament or hook proteins, the cell body rotates rapidly
about the stationary flagellum. If polystyrene-latex beads are attached to
flagella, the beads spin about the flagellar axis due to flagellar rotation.
The flagellar motor can rotate very rapidly.
The E.
coli motor rotates 270 revolutions per second; Vibrio alginolyticus averages
1,100 rps. Eucaryotic flagella and motility.
The
direction of flagellar rotation determines the nature of bacterial movement.
Monotrichous, polar flagella rotate counterclockwise (when viewed from outside
the cell) during normal forward movement, whereas the cell itself rotates
slowly clockwise.
The rotating helical flagellar filament thrusts the cell forward in a run with the flagellum trailing behind (figure 3.35). Monotrichous bacteria stop and tumble randomly by reversing the direction of flagellar rotation. Peritrichously flagellated bacteria operate in a somewhat similar way. To move forward, the flagella rotate counterclockwise. As they do so, they bend at their hooks to form a rotating bundle that propels them forward. Clockwise rotation of the flagella disrupts the bundle and the cell tumbles.
Because
bacteria swim through rotation of their rigid flagella, there must be some sort
of motor at the base. A rod or shaft extends from the hook and ends in the M
ring, which can rotate freely in the plasma membrane (figure 3.36). It
is believed that the S ring is attached to the cell wall in gram-positive cells
and does not rotate.
The P
and L rings of gram-negative bacteria would act as bearings for the rotating
rod. There is some evidence that the basal body is a passive structure and
rotates within a membrane-embedded protein complex much like the rotor of an
electrical motor turns in the center of a ring of electromagnets (the stator).
The exact
mechanism that drives basal body rotation still is not clear. Figure 3.36
provides a more detailed depiction of the basal body in gram-negative bacteria.
The rotor portion of the motor seems to be made primarily of a rod, the M ring,
and a C ring joined to it on the cytoplasmic side of the basal body. These two
rings are made of several proteins; Fli G is particularly important in
generating flagellar rotation. The two most important proteins in the stator part
of the motor are Mot A and Mot B. These form a proton channel through the
plasma membrane, and Mot B also anchors the Mot complex to cell wall
peptidoglycan. There is some evidence that Mot A and Fli G directly interact
during flagellar rotation. This rotation is driven by proton or sodium gradients
in procaryotes, not directly by ATP as is the case with eucaryotic flagella.
The
flagellum is a very effective swimming device. From the bacterium’s point of
view, swimming is quite a task because the surrounding water seems as thick and
viscous as molasses. The cell must bore through the water with its helical or
corkscrew shaped flagella, and if flagellar activity ceases, it stops almost
instantly.
Despite
such environmental resistance to movement, bacteria can swim from 20 to almost
90 µm/second. This is equivalent to traveling from 2 to over 100 cell lengths
per second.
In
contrast, an exceptionally fast 6 ft human might be able to run around 5 body
lengths per second. Bacteria can move by mechanisms other than flagellar
rotation. Spirochetes are helical bacteria that travel through viscous substances
such as mucus or mud by flexing and spinning movements caused by a special axial
filament composed of periplasmic flagella. A very different type of
motility, gliding motility, is employed by many bacteria: cyanobacteria,
myxobacteria and cytophagas, and some mycoplasmas.
Although
there are no visible external structures associated with gliding motility,
these bacteria can coast along solid surfaces at rates up to 3 µm/second.
3.7 Bacterial Chemotaxis
Bacteria do not always
swim aimlessly but are attracted by such nutrients as sugars and amino acids,
and are repelled by many harmful substances and bacterial waste products.
(Bacteria also can respond to other environmental cues such as temperature, light, and gravity; Box 3.3.) Movement toward
chemical attractants and away from repellents is known as chemotaxis. Such
behavior is of obvious advantage to bacteria.
Chemotaxis may be demonstrated by observing bacteria in the chemical gradient produced when a thin capillary tube is filled with an attractant and lowered into a bacterial suspension. As the attractant diffuses from the end of the capillary, bacteria collect and swim up the tube. The number of bacteria within the capillary after a short length of time reflects the strength of attraction and rate of chemotaxis.
Positive and
negative chemotaxis also can be studied with petri dish cultures (figure 3.37).
If bacteria are placed in the center of a dish of agar containing an
attractant, the bacteria will exhaust the local supply and then swim outward
following the attractant gradient they have created. The result is an expanding
ring of bacteria. When a disk of repellent is placed in a petri dish of
semisolid agar and bacteria, the bacteria will swim away from the repellent,
creating a clear zone around the disk (figure 3.38).
Bacteria can respond to very low
levels of attractants (about 10-8 M for some sugars), the magnitude
of their response increasing with attractant concentration. Usually they sense
repellents only at higher concentrations. If an attractant and a repellent are
present together, the bacterium will compare both signals and respond to the
chemical with the most effective concentration.
Attractants and repellents are detected by chemoreceptors, special proteins that bind chemicals and transmit signals to the other components of the chemosensing system. About 20 attractant chemoreceptors and 10 chemoreceptors for repellents have been discovered thus far. These chemoreceptor proteins may be located in the periplasmic space or the plasma membrane. Some receptors participate in the initial stages of sugar transport into the cell.
The chemotactic behavior of bacteria has been studied
using the tracking microscope, a microscope with a moving stage that automatically
keeps an individual bacterium in view. In the absence of a chemical gradient, E.
coli and other bacteria move randomly.
 |
A bacterium travels in a straight or
slightly curved line, a run, for a few seconds; then it will stop and tumble or
twiddle about. The tumble is followed by a run in a different direction (figure
3.39). When the bacterium is exposed to an attractant gradient, it tumbles less
frequently (or has longer runs) when traveling up the gradient, but tumbles at
normal frequency if moving down the gradient. Consequently the bacterium moves
up the gradient.
Behavior is shaped by temporal changes
in chemical concentration: the bacterium compares its current environment with that
experienced a few moments previously; if the attractant concentration is
higher, tumbling is suppressed and the run is longer.
The opposite response occurs with a
repellent gradient. Tumbling frequency decreases (the run lengthens) when the
bacterium moves down the gradient away from the repellent.
Although bacterial chemotaxis appears
to be deliberate, directed movement, it is important to keep in mind that this
is not the case. When the environment is constant, bacteria tend to move in a
random walk. That is, there is a random sequence of runs followed by tumbles.
If a run is in the direction of improving conditions, tumbles are suppressed so
that the cell tends to move in the preferred direction. This is said to be a
biased random walk toward attractants and away from repellants. Individual
cells do not choose a particular direction. Instead, they determine whether or not
to continue in the same direction.
Much work has been done on the mechanism
of chemotaxis in Escherichia coli. Recall that forward swimming is due to
counterclockwise rotation of the flagellum, whereas tumbling results from
clockwise rotation. The bacteria must be able to respond to gradients in such a
way that they collect in nutrient-rich regions and at the proper oxygen level
while avoiding toxic materials. E. coli has four different
chemoreceptors that recognize serine, aspartate and maltose, ribose and galactose,
and dipeptides, respectively. These chemoreceptors often are called
methyl-accepting chemotaxis proteins (MCPs).
They seem to be localized in patches,
often at the end of a rodshaped cell like E. coli. The MCPs do not
directly influence flagellar rotation but act through a series of proteins. The
whole process is so efficient that a stimulus can trigger a motor response in
less than 200 milliseconds.
The molecular mechanism underlying chemotaxis is quite complex. It involves conformational changes in proteins, protein methylation, and protein phosphorylation. When an attractant such as a nutrient molecule is not bound to an MCP, the CheA protein is phosphorylated using ATP. This phosphorylated protein can donate its phosphate to the CheY protein, which then interacts with the FliM switch protein at the base of the flagellum to promote clockwise rotation and tumbling.
An increase in nutrient
binding will lead to dephosphorylated CheA, counterclockwise flagellar
rotation, and a run. When no attractants or repellants are present, the system
maintains intermediate levels of CheA phosphate and CheY phosphate.
This produces a normal random walk
pattern. In very general terms, the system has a sensory protein that can be
phosphorylated and then phosphorylate another protein to cause a response.
As we shall see later, this is called
a two-component phosphorelay system. The molecular details of the chemotaxis system
will be described in chapter 12 when two-component systems are discussed. It
should be noted here that a similar mechanism is used to respond to other
environmental factors such as oxygen (aerotaxis), light (phototaxis),
temperature (thermotaxis), and osmotic pressure (osmotaxis).
3.8 The Bacterial Endospore
A number of gram-positive bacteria can form a special resistant, dormant structure called an endospore. Endospores develop within vegetative bacterial cells of several genera: Bacillus and Clostridium (rods), Sporosarcina (cocci), and others. These structures are extraordinarily resistant to environmental stresses such as heat, ultraviolet radiation, gamma radiation, chemical disinfectants, and desiccation.
In fact,
some endospores have remained viable for around 100,000 years, and actinomycete
spores (which are not true endospores) have been recovered alive after burial
in the mud for 7,500 years. Because of their resistance and the fact that
several species of endospore-forming bacteria are dangerous pathogens,
endospores are of great practical importance in food, industrial, and medical
microbiology. This is because it is essential to be able to sterilize solutions
and solid objects.
Endospores often survive
boiling for an hour or more; therefore autoclaves must be used to sterilize many
materials. Endospores are also of considerable theoretical interest. Because
bacteria manufacture these intricate entities in a very organized fashion over
a period of a few hours, spore formation is well suited for research on the
construction of complex biological structures. In the environment, endospores
aid in survival when moisture or nutrients are scarce.
Endospores can be examined with both light and electron microscopes. Because spores are impermeable to most stains, they often are seen as colorless areas in bacteria treated with methylene blue and other simple stains; special spore stains are used to make them clearly visible. Spore position in the mother cell or sporangium frequently differs among species, making it of considerable value in identification. Spores may be centrally located, close to one end (subterminal), or definitely terminal (figure 3.40). Sometimes a spore is so large that it swells the sporangium.
Electron
micrographs show that endospore structure is complex (figure 3.41). The spore
often is surrounded by a thin, delicate covering called the exosporium. A spore
coat lies beneath the exosporium, is composed of several protein layers, and
may be fairly thick. It is impermeable and responsible for the spore’s
resistance to chemicals. The cortex, which may occupy as much as half the spore
volume, rests beneath the spore coat. It is made of a peptidoglycan that is
less cross-linked than that in vegetative cells. The spore cell wall (or core
wall) is inside the cortex and surrounds the protoplast or core. The core has
the normal cell structures such as ribosomes and a nucleoid, but is
metabolically inactive.
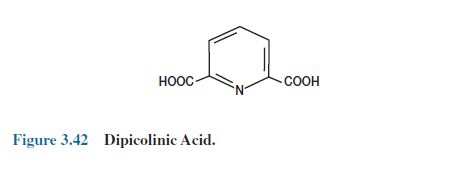
It is still not known precisely why the endospore is so resistant to heat and other lethal agents. As much as 15% of the spore’s dry weight consists of dipicolinic acid complexed with calcium ions (figure 3.42), which is located in the core. It has long been thought that dipicolinic acid was directly involved in spore heat resistance, but heat-resistant mutants lacking dipicolinic acid now have been isolated.
Calcium does aid in resistance to wet heat, oxidizing agents, and sometimes dry heat. It may be that calcium-dipicolinate often stabilizes spore nucleic acids. Recently specialized small, acid-soluble DNA-binding proteins have been discovered in the endospore.
They saturate spore DNA and protect it from heat, radiation, dessication, and chemicals. Dehydration of the protoplast appears to be very important in heat resistance. The cortex may osmotically remove water from the protoplast, thereby protecting it from both heat and radiation damage. The spore coat also seems to protect against enzymes and chemicals such as hydrogen peroxide. Finally, spores contain some DNA repair enzymes.
DNA is
repaired during germination and outgrowth after the core has become active once
again. In summary, endospore heat resistance probably is due to several
factors: calcium-dipicolinate and acid-soluble protein stabilization of DNA,
protoplast dehydration, the spore coat, DNA repair, the greater stability of
cell proteins in bacteria adapted to growth at high temperatures, and others.
Spore formation, sporogenesis or sporulation, normally commences when growth ceases due to lack of nutrients. It is a complex process and may be divided into seven stages (figure 3.43). An axial filament of nuclear material forms (stage I), followed by an inward folding of the cell membrane to enclose part of the DNA and produce the forespore septum (stage II).
The membrane continues to grow and engulfs the
immature spore in a second membrane (stage III). Next, cortex is laid down in
the space between the two membranes, and both calcium and dipicolinic acid are
accumulated (stage IV). Protein coats then are formed around the cortex (stage V),
and maturation of the spore occurs (stage VI). Finally, lytic enzymes destroy
the sporangium releasing the spore (stage VII). Sporulation requires only about
10 hours in Bacillus megaterium.
The transformation of
dormant spores into active vegetative cells seems almost as complex a process
as sporogenesis.
It occurs in three stages:
(1) activation, (2) germination, and (3) outgrowth. Often an endospore will not
germinate successfully, even in a nutrient-rich medium, unless it has been
activated.
Activation is a reversible process that prepares spores for germination and usually results from treatments like heating. It is followed by germination, the breaking of the spore’s dormant state. This process is characterized by spore swelling, rupture or absorption of the spore coat, loss of resistance to heat and other stresses, loss of refractility, release of spore components, and increase in metabolic activity.
Many normal metabolites or nutrients (e.g.,
amino acids and sugars) can trigger germination after activation. Germination
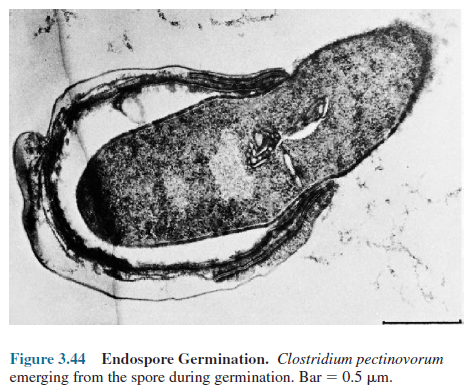
is followed by the third stage, outgrowth. The spore protoplast makes new components,
emerges from the remains of the spore coat, and develops again into an active
bacterium (figure 3.44).


































No comments:
Post a Comment