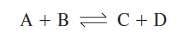Energy and Work
Energy may be most simply
defined as the capacity to do work or to cause particular changes. Thus all
physical and chemical processes are the result of the application or movement
of energy. Living cells carry out three major types of work, and all are
essential to life processes. Chemical work involves the synthesis of complex
biological molecules required by cells from much simpler precursors; energy is
needed to increase the molecular complexity of a cell.
Molecules and ions often must
be transported across cell membranes against an electro-chemical gradient. For
example, a molecule sometimes moves into a cell even though its concentration
is higher internally. Similarly a solute may be expelled from the cell against
a concentration gradient. This process is transport work and requires energy input
in order to take up nutrients, eliminate wastes, and maintain ion balances.
The third type of work is
mechanical work, perhaps the most familiar of the three. Energy is required to change
the physical location of organisms, cells, and structures within cells.
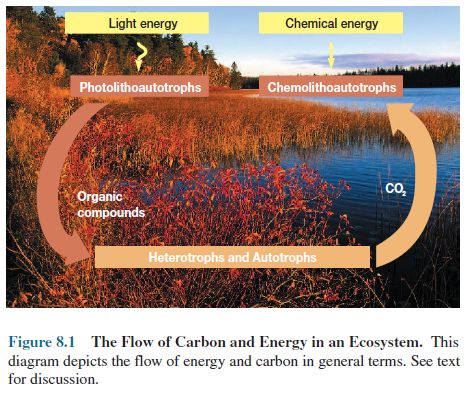
The ultimate source of
most biological energy is the visible sunlight impinging on the earth’s
surface. Light energy is trapped by phototrophs during photosynthesis, in which
it is absorbed by chlorophyll and other pigments and converted to chemical
energy. As noted in chapter 5, chemolithoautotrophs derive energy by oxidizing
inorganic compounds rather than obtaining it from light absorption.
Chemical energy from
photosynthesis and chemolithotrophy can then be used by photolithoautotrophs and
chemolithoautotrophs to transform CO2 into biological molecules such
as glucose (figure 8.1).
The complex molecules
manufactured by autotrophic organisms (both plant and microbial producers) serve
as a carbon source for chemoheterotrophs and other consumers that use complex organic
molecules as a source of material and energy for building their own cellular
structures (it should be remembered that autotrophs also use complex organic
molecules).
Chemoheterotrophs often
employ O2 as an electron acceptor when oxidizing glucose and other
organic molecules to CO2. This process, in which O2 acts
as the final electron acceptor and is reduced to water, is called aerobic
respiration. Much energy is released during
this process. Thus, in the ecosystem, energy is trapped by photoautotrophs and
chemolithoautotrophs; some of this energy subsequently flows to
chemoheterotrophs when they use nutrients derived from autotrophs (figure 8.1).
The CO2 produced during aerobic respiration
can be incorporated again into complex organic molecules during photosynthesis
and chemolithoautotrophy. Clearly the flow of carbon and energy in the
ecosystem is intimately related.
Cells must efficiently transfer energy
from their energygenerating or trapping apparatus to the systems actually
carrying out work. That is, cells must have a practical form of energy
currency.
In living organisms the major currency
is adenosine 5′- triphosphate (ATP; figure 8.2). When ATP breaks down to adenosine
diphosphate (ADP) and orthophosphate (Pi), energy is made available
for useful work. Later, energy from photosynthesis, aerobic respiration,
anaerobic respiration, and fermentation is used to resynthesize ATP from ADP
and Pi. An energy cycle is created in the cell (figure 8.3).
The Laws of Thermodynamics
To understand how energy is trapped or generated and how ATP functions as an energy currency, some knowledge of the basic principles of thermodynamics is required. The science of thermodynamics analyzes energy changes in a collection of matter (e.g., a cell or a plant) called a system. All other matter in the universe is called the surroundings. Thermodynamics focuses on the energy differences between the initial state and the final state of a system. It is not concerned with the rate of the process.
For instance, if a pan of water is heated to boiling, only
the condition of the water at the start and at boiling is important in
thermodynamics, not how fast it is heated or on what kind of stove. Two
important laws of thermodynamics must be understood. The first law of
thermodynamics says that energy can be neither created nor destroyed. The total
energy in the universe remains constant although it can be redistributed. For
example, many energy exchanges do occur during chemical reactions (e.g., heat
is given off by exothermic reactions and absorbed during endothermic
reactions), but these heat exchanges do not violate the first law.
It is necessary to specify quantitatively the amount of energy used in or evolving from a particular process, and two types of energy units are employed. A calorie (cal) is the amount of heat energy needed to raise one gram of water from 14.5 to 15.5°C. The amount of energy also may be expressed in terms of joules (J), the units of work capable of being done.
One cal of heat is equivalent to 4.1840 J of work.
One thousand calories or a kilocalorie (kcal) is enough energy to boil 1.9 ml
of water. A kilojoule is enough energy to boil about 0.44 ml of water, or
enable a person weighing 70 kg to climb 35 steps. The joule is normally used by
chemists and physicists. Because biologists most often speak of energy in terms
of calories, this text will employ calories when discussing energy changes.
 |
Although it is true that
energy is conserved in the universe, the first law of thermodynamics does not
account for many physical and chemical processes. A simple example may help
make this clear. Suppose a full gas cylinder is connected to an empty one by a
tube with a valve (figure 8.4). If the valve is opened, gas flows from the full
to the empty cylinder until the gas pressure is equal on both sides. Energy has
not only been redistributed but also conserved.
The expansion of gas is
explained by the second law of thermodynamics and a condition of matter called
entropy. Entropy may be considered a measure of the randomness or disorder of a
system. The greater the disorder of a system, the greater is its entropy. The
second law states that physical and chemical processes proceed in such a way
that the randomness or disorder of the universe (the system and its surroundings)
increases to the maximum possible. Gas will always expand into an empty
cylinder.
Free Energy and Reactions
The first and second laws
can be combined in a useful equation, relating the changes in energy that can
occur in chemical reactions and other processes.
ΔG = ΔH – T x
ΔS
ΔG is the change in free energy, ΔH is the change in enthalpy, T is the temperature in Kelvin (°C -273), and ΔS is the change in entropy occurring during the reaction. The change in enthalpy is the change in heat content. Cellular reactions occur under conditions of constant pressure and volume. Thus the change in enthalpy is about the same as the change in total energy during the reaction. The free energy change is the amount of energy in a system available to do useful work at constant temperature and pressure.
Therefore the change in entropy is a measure of the
proportion of the total energy change that the system cannot use in performing
work. Free energy and entropy changes do not depend on how the system gets from
start to finish. A reaction will occur spontaneously at constant temperature
and pressure if the free energy of the system decreases during the reaction or,
in other words, if ΔG is negative. It follows from the equation that a reaction
with a large positive change in entropy will normally tend to have a negative ΔG
value and therefore occur spontaneously.
A decrease in entropy will
tend to make ΔG more positive and the reaction less favorable.
The change in free energy has a definite,
concrete relationship to the direction of chemical reactions. Consider the
following simple reaction:
![]()
If the molecules A and B are mixed,
they will combine to form the products C and D. Eventually C and D will become
concentrated enough to combine and produce A and B at the same rate as they are
formed from A and B. The reaction is now at equilibrium: the rates in both
directions are equal and no further net change occurs in the concentrations of
reactants and products.
This situation is described by the
equilibrium constant (Keq), relating the equilibrium concentrations of
products and substrates to one another.
Keq =[C][D]/[A][B]
If the equilibrium constant is greater
than one, the products are in greater concentration than the reactants at
equilibrium—that is, the reaction tends to go to completion as written.
The equilibrium constant of a reaction
is directly related to its change in free energy. When the free energy change
for a process is determined at carefully defined standard conditions of
concentration, pressure, pH, and temperature, it is called the standard free
energy change (ΔGo). If the pH is set at 7.0 (which is close
to the pH of living cells), the standard free energy change is indicated by the
symbol ΔGo′.
The change in standard free energy may
be thought of as the maximum amount of energy available from the system for
useful work under standard conditions. Using ΔGo′ values allows one to compare
reactions without worrying about variations in the ΔG due to differences
in environmental conditions. The relationship between ΔGo′ and Keq
is given by the following equation:
ΔGo´= -2.303RT.logKeq
R is the gas constant (1.9872 cal/mole-degree or
8.3145 J/moledegree), and T is the absolute temperature. Inspection of
this equation shows that when ΔGo′ is negative, the
equilibrium constant is greater than one and the reaction goes to completion as
written. It is said to be an exergonic reaction (figure 8.5). In an endergonic reaction
ΔGo′ is positive and the equilibrium constant is less than one.
That is, the reaction is not favorable, and little product will be formed at equilibrium
under standard conditions. Keep in mind that the ΔGo′ value
shows only where the reaction lies at equilibrium, not how fast the reaction
reaches equilibrium.
The Role of ATP in Metabolism
Many reactions in the cell
are endergonic and will not proceed far toward completion without outside
assistance. One of ATP’s major roles is to drive such endergonic reactions more
to completion.
ATP is a high-energy
molecule. That is, it breaks down or hydrolyzes almost completely to the
products ADP and Pi with a ΔGo′ of _7.3 kcal/mole.
ATP + H2O > ADP + Pi
In reference to ATP the
term high-energy molecule does not mean that there is a great deal of energy
stored in a particular bond of ATP. It simply indicates that the removal of the
terminal phosphate goes to completion with a large negative standard free
energy change, or the reaction is strongly exergonic. In other words, ATP has a
high phosphate group transfer potential; it readily transfers its phosphate to
water. The phosphate group transfer potential is defined as the negative of ΔGo′ for the
hydrolytic removal of phosphate. A molecule with a higher group transfer
potential will donate phosphate to one with a lower potential.
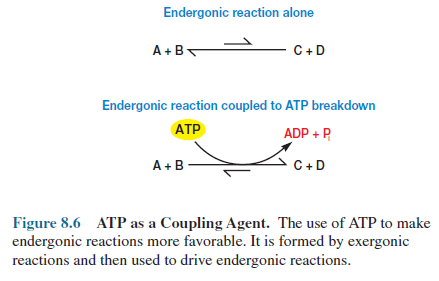
Thus ATP is ideally suited
for its role as an energy currency.
It is formed in
energy-trapping and -generating processes such as photosynthesis, fermentation,
and aerobic respiration. In the cell’s economy, exergonic ATP breakdown is coupled
with various endergonic reactions to promote their completion (figure 8.6). In other
words ATP links energy-generating reactions, which liberate free energy, with
energy-using reactions, which require free energy input to proceed toward
completion. Facilitation of chemical work is the focus of the preceding
example, but the same principles apply when ATP is coupled with endergonic
processes involving transport work and mechanical work (figure 8.3).
Oxidation-Reduction Reactions and Electron
Carriers
Free energy changes are
not only related to the equilibria of “regular” chemical reactions but also to
the equilibria of oxidationreduction reactions. The release of energy normally
involves oxidation-reduction reactions. Oxidation-reduction (redox) reactions are
those in which electrons move from a donor, the reducing agent or reductant, to
an electron acceptor, the oxidizing
agent or oxidant. By convention such a reaction is written with the reductant
to the right of the oxidant and the number (n) of electrons (e_)
transferred.
Oxidant
+ ne– > reductant
The oxidant and reductant pair is
referred to as a redox couple (table 8.1). When an oxidant accepts electrons,
it becomes the reductant of the couple. The equilibrium constant for the
reaction is called the standard reduction potential (E0) and
is a measure of the tendency of the reducing agent to lose electrons. The
reference standard for reduction potentials is the hydrogen system with an E′0
(the reduction potential at pH 7.0) of _0.42 volts or _420
millivolts.
2H+
+ 2e– > H2
In this reaction each hydrogen atom
provides one proton (H+) and one electron (e_).
The reduction potential has a concrete
meaning. Redox couple with more negative reduction potentials will donate
electrons to couples with more positive potentials and greater affinity for electrons.
Thus electrons will tend to move from reductants at the top of the list in
table 8.1 to oxidants at the bottom because they have more positive potentials.
This may be expressed visually in the
form of an electron tower in which the most negative reduction potentials are
at the top (figure 8.7). Electrons move from donors to acceptors down the
potential gradient or fall down the tower to more positive potentials. Consider
the case of the electron carrier nicotinamide adenine dinucleotide (NAD+).
The NAD+/NADH couple has a very negative E′0 and
can therefore give electrons to many acceptors, including O2.
Because NAD+/NADH is more
negative than 1/2 O2/H2O, electrons will flow from NADH
(the reductant) to O2 (the oxidant) as shown in figure 8.7.
![]() NADH
+ H+ + 1/2O2 > H2O + NAD+
NADH
+ H+ + 1/2O2 > H2O + NAD+
When electrons move from a reductant
to an acceptor with a more positive redox potential, free energy is released.
The ΔGo′ of the reaction is directly related to the magnitude
of the difference between the reduction potentials of the two couples (ΔE′0).
The larger the ΔE′0, the greater the amount of free energy made
available, as is evident from the equation
ΔGo´=__nF.ΔE´0
in which n is the number of
electrons transferred and F is the Faraday constant (23,062
cal/mole-volt or 96,494 J/mole-volt).
For every 0.1 volt change in ΔE′0,
there is a corresponding 4.6 kcal change in ΔGo′ when a two-electron
transfer takes place. This is similar to the relationship of ΔGo′ and Keq
in other chemical reactions—the larger the equilibrium constant, the greater the
ΔGo′. The difference in reduction potentials between NAD+/NADH
and 1/2O2/H2O is 1.14 volts, a large ΔE′0
value.
When electrons move from NADH to O2
during aerobic respiration, a large amount of free energy is made available to
synthesize ATP (figure 8.8). If energy is released when electrons flow from
negative to positive reduction potentials, then an input of energy is required
to move electrons in the opposite direction, from more positive to more
negative potentials. This is precisely what occurs during photosynthesis
(figure 8.8). Light energy is trapped and used to move electrons from water to
the electron carrier nicotinamide adenine dinucleotide phosphate (NADP+).
The cycle of energy flow discussed
earlier and illustrated in figure 8.1 can be understood from a different
perspective, if the preceding concept is kept in mind. Photosynthetic organisms
capture light energy and use it to move electrons from water (and other
electron donors such as H2S) to electron acceptors, such as NADP+, that have more negative reduction
potentials. These electrons can then flow back to more positive acceptors and
provide energy for ATP production during photosynthesis. Photoautotrophs use
ATP and NADPH to synthesize complex molecules from CO2.
Chemoheterotrophs also make use of energy released during the movement of
electrons by oxidizing complex nutrients during respiration to produce NADH.
NADH subsequently donates its electrons to O2, and the energy
released during electron transfer is trapped in the form of ATP. The energy
from sunlight is made available to all living organisms because of this
relationship between electron flow and energy.
Electron transport is
important in aerobic respiration, anaerobic respiration, chemolithotrophy, and
photosynthesis. Electron movement in cells requires the participation of
carriers such as NAD+ and NADP+, both of which can
transport electrons between different locations. The nicotinamide ring of NAD+
and NADP+ (figure 8.9) accepts two electrons and one proton from a donor,
while a second proton is released. There are several other electron carriers of
importance in microbial metabolism (table 8.1), and they carry electrons in a
variety of ways.
Flavin adenine dinucleotide
(FAD) and flavin mononucleotide (FMN) bear two electrons and two protons on the
complex ring system shown in figure 8.10. Proteins bearing FAD and FMN are
often called flavoproteins. Coenzyme Q (CoQ) or ubiquinone is a quinone that
transports electrons and protons in many respiratory electron transport chains
(figure 8.11). Cytochromes and several other carriers
use iron atoms to transport electrons by reversible oxidation and reduction
reactions.
Fe3+
(ferric iron) + e– > Fe2+
(ferrous iron)
In the cytochromes these iron atoms
are part of a heme group (figure 8.12) or other similar iron-porphyrin rings.
Several different cytochromes, each of which consists of a protein and an
iron porphyrin ring, are a prominent part of respiratory electron transport chains.
Some iron containing electron-carrying proteins lack a heme group and are
called nonheme iron proteins. Ferredoxin is a nonheme iron protein active in
photosynthetic electron transport and several other electron transport
processes.
Even though its iron atoms are not bound to a heme group, they still undergo reversible oxidation and reduction reactions. Although all the previously discussed molecules function in electron transport chains, some bear two electrons (NAD, FAD, and CoQ) while others carry only one electron at a time (cytochromes and nonheme iron proteins). This difference in the number of electrons carried is of great importance in the operation of electron transport chains.
Enzymes
Recall that an exergonic
reaction is one with a negative ΔGo′ and an equilibrium
constant greater than one. An exergonic reaction will proceed to completion in
the direction written (that is, toward the right of the equation).
Nevertheless, one often can combine the reactants for an exergonic reaction
with no obvious result, even though products should be formed. It is precisely
in these reactions that enzymes play their part.
Structure and Classification of
Enzymes
Enzymes may be defined as
protein catalysts that have great specificity for the reaction catalyzed and
the molecules acted on. A catalyst is a substance that increases the rate of a
chemical reaction without being permanently altered itself. Thus enzymes speed
up cellular reactions. The reacting molecules are called substrates, and the
substances formed are the products.
Many enzymes are indeed
pure proteins. However, many enzymes consist of a protein, the apoenzyme, and
also a nonprotein component, a cofactor, required for catalytic activity.
The complete enzyme
consisting of the apoenzyme and its cofactor is called the holoenzyme. If the
cofactor is firmly attached to the apoenzyme it is a prosthetic group. Often the
cofactor is loosely attached to the apoenzyme. It can even dissociate from the
enzyme protein after products have been formed and carry one of these products
to another enzyme (figure 8.13). Such a loosely bound cofactor is called a
coenzyme.
For example, NAD+
is a coenzyme that carries electrons within the cell. Many vitamins that humans
require serve as coenzymes or as their precursors. Niacin is incorporated into
NAD+ and riboflavin into FAD. Metal ions may also be bound to
apoenzymes and act as cofactors.
Despite the large number
and bewildering diversity of enzymes present in cells, they may be placed in
one of six general classes (table 8.2). Enzymes usually are named in terms of
the substrates they act on and the type of reaction catalyzed. For example, lactate
dehydrogenase (LDH) removes hydrogens from lactate.
Lactate + NAD+ LDH > pyruvate + NADH + H+
Lactate dehydrogenase can
also be given a more complete and detailed name, L-lactate: NAD oxidoreductase.
This name describes the substrates and reaction type with even more precision.
The Mechanism of Enzyme Reactions
It is important to keep in
mind that enzymes increase the rates of reactions but do not alter their
equilibrium constants. If a reaction is endergonic, the presence of an enzyme
will not shift its equilibrium so that more
products can be formed. Enzymes simply speed up the rate
at which a reaction proceeds toward its final equilibrium.
How do enzymes catalyze reactions?
Although a complete answer would be long and complex, some understanding of the
mechanism can be gained by considering the course of a normal exergonic
chemical reaction.
![]() A
+ B C + D
A
+ B C + D
When molecules A and B approach each
other to react, they form a transition-state complex, which resembles both the
substrates and the products (figure 8.14). The activation energy is required to
bring the reacting molecules together in the correct way to reach the transition
state. The transition-state complex can then decompose to yield the products C
and D. The difference in free energy level between reactants and products is ΔGo′.
Thus the equilibrium in our example will lie toward the products because ΔGo′
is negative (i.e., the products are at a lower energy level than the substrates).
Clearly A and B will not be converted
to C and D in figure 8.14 if they are not supplied with an amount of energy
equivalent to the activation energy. Enzymes accelerate reactions by lowering the
activation energy; therefore more substrate molecules will have sufficient
energy to come together and form products. Even though the equilibrium constant
(or ΔGo′) is unchanged, equilibrium will be reached more rapidly in the
presence of an enzyme because of this decrease in the activation energy.
Researchers have expended much effort
in discovering how enzymes lower the activation energy of reactions, and the
process is becoming clearer. Enzymes bring substrates together at a special place
on their surface called the active site or catalytic site to form an
enzyme-substrate complex (figures 8.15, 8.16). The enzyme can interact with a
substrate in two general ways. It may be rigid and shaped to precisely fit the
substrate so that the correct substrate binds specifically and is positioned
properly for reaction. This mechanism is referred to as the lock-and-key model.
An enzyme also may change shape when it binds the substrate so that the active
site surrounds and precisely fits the substrate.
This has been called the induced fit
model and is used by hexokinase and many other enzymes (figure 8.16). The
formation of an enzyme-substrate complex can lower the activation energy in many
ways. For example, by bringing the substrates together at the active site, the
enzyme is, in effect, concentrating them and speeding up the reaction. An
enzyme does not simply concentrate its substrates, however.
It also binds them so that they are
correctly oriented with respect to each other in order to form a
transition-state complex. Such an orientation lowers the amount of energy that
the substrates require to reach the transition state. These and other catalytic
site activities speed up a reaction hundreds of thousands of times, even though
microorganisms grow between _20°C and approximately 113°C. These
temperatures are not high enough to favor most organic reactions in the absence
of enzyme catalysis, yet cells cannot survive at the high temperatures used by
an organic chemist in routine organic syntheses. Enzymes make life possible by
accelerating specific reactions at low temperatures.
The Effect of Environment on Enzyme Activity
Enzyme activity varies greatly with
changes in environmental factors, one of the most important being the substrate
concentration.
As will be emphasized later, substrate
concentrations are usually low within cells. At very low substrate
concentrations, an enzyme makes product slowly because it seldom contacts a
substrate molecule. If more substrate molecules are present, an enzyme binds
substrate more often, and the reaction velocity (usually expressed in terms of
the rate of product formation) is greater than at a lower substrate
concentration. Thus the rate of an enzyme-catalyzed reaction increases with
substrate concentration (figure 8.17).
Eventually further increases in
substrate concentration do not result in a greater reaction velocity because
the available enzyme molecules are binding substrate and converting it to
product as rapidly as possible. That is, the enzyme is saturated with substrate
and operating at maximal velocity (Vmax).
The resulting substrate concentration
curve is a hyperbola (figure 8.17). It is useful to know the substrate
concentration an enzyme needs to function adequately. Usually the Michaelis
constant (Km), the substrate concentration required for the enzyme to achieve
half maximal velocity, is used as a measure of the apparent affinity of an
enzyme for its substrate. The lower the Km value, the lower the
substrate concentration at which an enzyme catalyzes its reaction.
Enzymes also change activity with
alterations in pH and temperature (figure 8.18). Each enzyme functions most
rapidly at a specific pH optimum. When the pH deviates too greatly from an enzyme’s
optimum, activity slows and the enzyme may be damaged.
Enzymes likewise have temperature
optima for maximum activity. If the temperature rises too much above the
optimum, an enzyme’s structure will be disrupted and its activity lost. This phenomenon,
known as denaturation, may be caused by extremes of pH and temperature or by
other factors. The pH and temperature optima of a microorganism’s enzymes often
reflect the pH and temperature of its habitat. Not surprisingly bacteria growing
best at high temperatures often have enzymes with high temperature optima and
great heat stability.
Enzyme Inhibition
Microorganisms can be
poisoned by a variety of chemicals, and many of the most potent poisons are
enzyme inhibitors. A competitive inhibitor directly competes with the substrate
at an enzyme’s catalytic site and prevents the enzyme from forming product. A
classic example of this behavior is seen with the enzyme succinate
dehydrogenase, which catalyzes the oxidation of succinate to fumarate in the
tricarboxylic acid cycle. Malonic acid is an effective competitive inhibitor of
succinate dehydrogenase because it so closely resembles succinate, the normal
substrate (figure 8.19).
After malonate binds to
the enzyme, it cannot be oxidized and the formation of fumarate is blocked.
Competitive inhibitors usually resemble normal substrates, but they cannot be
converted to products.
Competitive inhibitors are
important in the treatment of many microbial diseases. Sulfa drugs like
sulfanilamide resemble p-aminobenzoate, a molecule used in the formation
of the coenzyme folic acid. The drugs compete with p-aminobenzoate for the
catalytic site of an enzyme involved in folic acid synthesis. This blocks the
production of folic acid and inhibits bacterial growth. Humans are not harmed
because they cannot synthesize folic acid and must obtain it in their diet.
Inhibitors also can affect
enzyme activity by binding to the enzyme at some location other than at the
active site. This alters the enzyme’s shape, rendering it inactive or less
active. These inhibitors are often called noncompetitive because they do not
directly compete with the substrate. Heavy metal poisons like mercury frequently
are noncompetitive inhibitors of enzymes.
The Nature and Significance of Metabolic
Regulation
The task of the regulatory
machinery is exceptionally complex and difficult. Pathways must be regulated
and coordinated so effectively that all cell components are present in
precisely the correct amounts. Furthermore, a microbial cell must be able to
respond effectively to environmental changes by using those nutrients present at
the moment and by switching on new catabolic pathways when different nutrients
become available. Because all chemical components of a cell usually are not
present in the surroundings, microorganisms also must synthesize unavailable
components and alter biosynthetic activity in response to changes in nutrient
availability.
The chemical composition
of a cell’s surroundings is constantly changing, and these regulatory processes
are dynamic and continuously responding to altered conditions.
Regulation is essential
for the cell to conserve microbial energy and material and to maintain
metabolic balance. If a particular energy source is unavailable, the enzymes
required for its use are not needed and their further synthesis is a waste of
carbon, nitrogen, and energy. Similarly it would be extremely wasteful for a
microorganism to synthesize the enzymes required to manufacture a certain end
product if that end product were already present in adequate amounts. Thus both
catabolism and anabolism are regulated in such a way as to maximize efficiency
of operation.
The drive to maintain balance and conserve energy and material is evident in the regulatory responses of a bacterium like E. coli. If the bacterium is grown in a very simple medium containing only glucose as a carbon and energy source, it will synthesize the required cell components in balanced amounts. Addition of a biosynthetic end product (the amino acid tryptophan, for example) to the medium will result in the immediate inhibition of the pathway synthesizing that end product; synthesis of the pathway’s enzymes also will slow or cease.
If E. coli is
transferred to medium containing only the sugar lactose, it will synthesize the
enzymes required for catabolism of this nutrient. In contrast, when E. coli grows
in a medium possessing both glucose and lactose, glucose (the sugar supporting
most rapid growth) is catabolized first. The culture will use lactose only
after the glucose supply has been exhausted.
The flow of carbon through
a pathway may be regulated in three major ways.
1. The localization of metabolites and enzymes in different parts of a cell, a phenomenon called metabolic channeling, influences pathway activity.
2. Critical enzymes often are directly stimulated or inhibited to alter pathway activity rapidly.
3. The number of enzyme molecules also may be controlled. The more catalyst molecules present, the greater the pathway’s activity. In bacteria regulation is usually exerted at the level of transcription. Control of mRNA synthesis is slower than direct regulation of enzyme activity but does result in the saving of much energy and raw material because enzymes are not synthesized when not required.
Each of these mechanisms
is described in detail. This chapter introduces the first two: metabolic channeling
and direct control of enzyme activity.
Metabolic Channeling
One of the most common channeling mechanisms is that of compartmentation, the differential distribution of enzymes and metabolites among separate cell structures or organelles. Compartmentation is particularly important in eucaryotic microorganisms with their many membrane-bound organelles. For example, fatty acid oxidation is located within the mitochondrion, whereas fatty acid synthesis occurs in the cytoplasmic matrix. The periplasm in procaryotes can also be considered an example of compartmentation.
Compartmentation makes possible the simultaneous, but
separate, operation and regulation of similar pathways. Furthermore, pathway
activities can be coordinated through regulation of the transport of
metabolities and coenzymes between cell compartments. Suppose two pathways in
different cell compartments require NAD for activity. The distribution of NAD
between the two compartments will then determine the relative activity of these
competing pathways, and the pathway with access to the most NAD will be
favored.
Channeling also occurs
within compartments such as the cytoplasmic matrix. The matrix is a structured
dense material with many subcompartments. In eucaryotes it also is subdivided
by the endoplasmic reticulum and cytoskeleton. Metabolites and coenzymes do not
diffuse rapidly in such an environment, and metabolite gradients will build up
near localized enzymes or enzyme systems. This occurs because enzymes at a
specific site convert their substrates to products, resulting in decreases in
the concentration of one or more metabolites and increases in others.
For example, product
concentrations will be high near an enzyme and decrease with increasing
distance from it.
Channeling can generate marked variations in metabolite concentrations and therefore directly affect enzyme activity. Substrate levels are generally around 10-3 moles/liter (M) to 10-6 M or even lower. Thus they may be in the same range as enzyme concentrations and equal to or less than the Michaelis constants (Km) of many enzymes. Under these conditions the concentration of an enzyme’s substrate may control its activity because the substrate concentration is in the rising portion of the hyperbolic substrate saturation curve (figure 8.20).
As the substrate level increases, it is converted to product more
rapidly; a decline in substrate concentration automatically leads to lower
enzyme activity. If two enzymes in different pathways use the same metabolite,
they may directly compete for it. The pathway winning this competition—the one
with the enzyme having the lowest Km value for the metabolite—will
operate closer to full capacity. Thus channeling within a cell compartment can
regulate and coordinate metabolism through variations in metabolite and coenzyme
levels.
Control of Enzyme Activity
Adjustment of the activity of regulatory enzymes controls the functioning of many metabolic pathways. This section describes these enzymes and discusses their role in regulating pathway activity.
Allosteric Regulation
Usually regulatory enzymes
are allosteric enzymes. The activity of an allosteric enzyme is altered by a
small molecule known as an effector or modulator. The effector binds reversibly
by noncovalent forces to a regulatory site separate from the catalytic site and
causes a change in the shape or conformation of the enzyme (figure 8.21). The
activity of the catalytic site is altered as a result. A positive effector
increases enzyme activity, whereas a negative effector decreases activity or
inhibits the enzyme. These changes in activity often result from alterations in
the apparent affinity of the enzyme for its substrate, but changes in maximum velocity
also can occur.
The kinetic characteristics
of nonregulatory enzymes show that the Michaelis constant (Km) is the
substrate concentration required for an enzyme to operate at half its maximal
velocity. This constant applies only to hyperbolic substrate saturation curves,
not to the sigmoidal curves often seen with allosteric enzymes (figure 8.23).
The substrate concentration required for half maximal velocity with allosteric
enzymes having sigmoidal substrate curves is called the [S]0.5 or K0.5
value.
One of the best-studied
allosteric regulatory enzymes is the aspartate carbamoyltransferase (ACTase)
from E. coli. The enzyme catalyzes the condensation of carbamoyl
phosphate with aspartate to form carbamoylaspartate (figure 8.22). ACTase
catalyzes the rate-determining reaction of the pyrimidine biosynthetic pathway
in E. coli. The substrate saturation curve is sigmoidal when the
concentration of either substrate is varied (figure 8.23). The enzyme has more
than one active site, and the binding of a substrate molecule to an active site
increases the binding of substrate at the other sites. In addition, cytidine triphosphate
(CTP), an end product of pyrimidine biosynthesis, inhibits the enzyme and the
purine ATP activates it. Both effectors alter the K0.5 value of the
enzyme but not its maximum velocity.
CTP inhibits by increasing
K0.5 or by shifting the substrate saturation curve to higher values.
This allows the enzyme to operate more slowly at a particular substrate
concentration when CTP is present. ATP activates by moving the curve to lower
substrate concentration values so that the enzyme is maximally active over a
wider substrate concentration range. Thus when the pathway is so active that
the CTP concentration rises too high, ACTase activity decreases and the rate of
end product formation slows. In contrast, when the purine end product ATP increases
in concentration relative to CTP, it stimulates CTP synthesis through its
effects on ACTase.
E.
coli aspartate carbamoyltransferase
provides a clear example of separate regulatory and catalytic sites in
allosteric enzymes. The enzyme is a large protein composed
of two catalytic subunits and three regulatory
subunits (figure 8.24a). The catalytic subunits contain only catalytic
sites and are unaffected by CTP and ATP. Regulatory subunits do not catalyze
the reaction but do possess regulatory sites to which CTP and ATP bind. When these
effectors bind to the complete enzyme, they cause conformational changes in the
regulatory subunits and subsequently in the catalytic subunits and their
catalytic sites. The enzyme can change reversibly between a less active T form
and a more active R form (figure 8.24b,c). Thus the regulatory site
influences a catalytic site about 6.0 nm distant.
Covalent Modification of Enzymes
Regulatory enzymes also can be switched on and off by reversible covalent modification. Usually this occurs through the addition and removal of a particular group, one form of the enzyme being more active than the other. For example, glycogen phosphorylase of the bread mold Neurospora crassa exists in phosphorylated and dephosphorylated forms called phosphorylase a and phosphorylase b, respectively (figure 8.25).
Phosphorylase b is inactive because
its required activator AMP is usually not present at sufficiently high levels. Phosphorylase a, the phosphorylated
form, is active even without AMP. Glycogen phosphorylase is stimulated by
phosphorylation of phosphorylase b to produce phosphorylase a. The
attachment of phosphate changes the enzyme’s conformation to an active form.
Phosphorylation and dephosphorylation are catalyzed by separate enzymes, which
also are regulated.
Enzymes can be regulated through
the attachment of groups other than phosphate. One of the most intensively
studied regulatory enzymes is E. coli glutamine synthetase, a large,
complex enzyme existing in two forms (figure 8.26). When an adenylic acid residue
is attached to each of its 12 subunits forming an adenylylated enzyme,
glutamine synthetase is not very active. Removal f AMP groups produces more
active deadenylylated glutamine synthetase, and glutamine is formed. The
glutamine synthetase system differs from the phosphorylase system in two ways: (1)
AMP is used as the modifying agent, and (2) the modified form of glutamine
synthetase is less active. Glutamine synthetase also is allosterically
regulated.
There are some advantages
to using covalent modification for the regulation of enzyme activity. These
interconvertible enzymes often are also allosteric. Because each form can
respond differently to allosteric effectors, systems of covalently modified enzymes
are able to respond to more stimuli in varied and sophisticated ways.
Regulation can also be exerted on the enzymes that catalyze the covalent
modifications, which adds a second level of regulation to the system.
Feedback Inhibition
The rate of many metabolic
pathways is adjusted through control of the activity of the regulatory enzymes
described in the preceding section. Every pathway has at least one pacemaker
enzyme that catalyzes the slowest or rate-limiting reaction in the pathway. Because
other reactions proceed more rapidly than the pacemaker reaction, changes in
the activity of this enzyme directly alter the speed with which a pathway
operates. Usually the first step in a pathway is a pacemaker reaction catalyzed
by a regulatory enzyme.
The end product of the
pathway often inhibits this regulatory enzyme, a process known as feedback
inhibition or end product inhibition. Feedback inhibition ensures balanced
production of a pathway end product. If the end product becomes too concentrated,
it inhibits the regulatory enzyme and slows its own synthesis. As the end
product concentration decreases, pathway activity again increases and more
product is formed. In this way feedback inhibition automatically matches end
product supply with the demand. The previously discussed E. coli aspartate
carbamoyl transferase is an excellent example of end product or feedback inhibition.
Frequently a biosynthetic pathway
branches to form more than one end product. In such a situation the synthesis
of pathway end products must be coordinated precisely. It would not do to have
one end product present in excess while another is lacking.
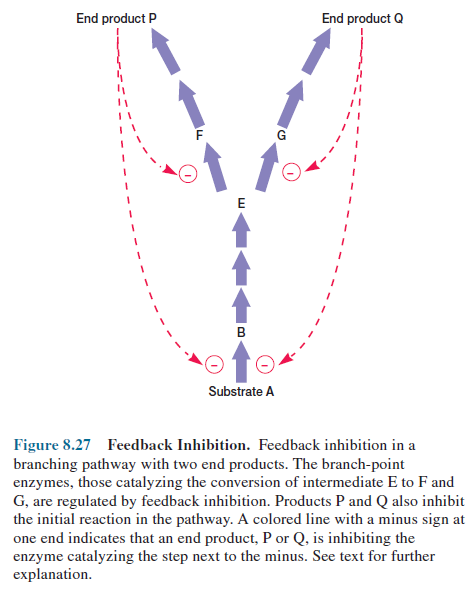
Branching biosynthetic pathways
usually achieve a balance between end products through the use of regulatory
enzymes at branch points (figure 8.27). If an end product is present in excess,
it often inhibits the branch-point enzyme on the sequence leading to its
formation, in this way regulating its own formation without affecting the
synthesis of other products. In figure 8.27 notice that both products also
inhibit the initial enzyme in the pathway. An excess of one product slows the
flow of carbon into the whole pathway while inhibiting the appropriate
branch-point enzyme. Because less carbon is required when a branch is not functioning,
feedback inhibition of the initial pacemaker enzyme helps match the supply with
the demand in branching pathways.
The regulation of multiple branched
pathways is often made even more sophisticated by the presence of isoenzymes,
different enzymes that catalyze the same reaction. The initial pacemaker step
may be catalyzed by several isoenzymes, each under separate and independent
control. In such a situation an excess of a single end product reduces pathway
activity but does not completely block pathway function because some isoenzymes
are still active.